Spectral Clustering
specClust.RdSpectral clustering based on k-nearest neighbor graph.
specClust(data, centers=NULL, nn = 7, method = "symmetric", gmax=NULL, ...) # S3 method for specClust plot(x, ...)
Arguments
| data | Matrix or data frame. |
|---|---|
| centers | number of clusters to estimate, if NULL the number is chosen automatical. |
| nn | Number of neighbors considered. |
| method | Normalisation of the Laplacian ("none", "symmetric" or "random-walk"). |
| gmax | maximal number of connected components. |
| x | an object of class |
| ... | Further arguments passed to or from other methods. |
Details
specClust alllows to estimate several popular spectral clustering algorithms, for an overview see von Luxburg (2007).
The Laplacian is constructed from a from nearest neighbors and there are several kernels available. The eigenvalues and eigenvectors are computed using the binding in igraph to arpack. This should ensure that this algorithm is also feasable for larger datasets as the the the distances used have dimension n*m, where n is the number of observations and m the number of nearest neighbors. The Laplacian is sparse and has roughly n*m elements and only k eigenvectors are computed, where k is the number of centers.
Value
specClust returns a kmeans object or in case of k being a vector a list of kmeans objects.
References
U. von Luxburg (2007) A tutorial on spectral clustering, Stat Comput, 17, 395--416
Ng, A., Jordan, M., Weiss, Y. (2002) On spectral clustering: analysis and an algorithm. In: Dietterich, T., Becker, S., Ghahramani, Z. (eds.) Advances in Neural Information Processing Systems, 14, 849--856. MIT Press, Cambridge
Lihi Zelnik-Manor and P. Perona (2004) Self-Tuning Spectral Clustering, Eighteenth Annual Conference on Neural Information Processing Systems, (NIPS)
Shi, J. and Malik, J. (2000). Normalized cuts and image segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence, 22 (8), 888--905
See also
Examples
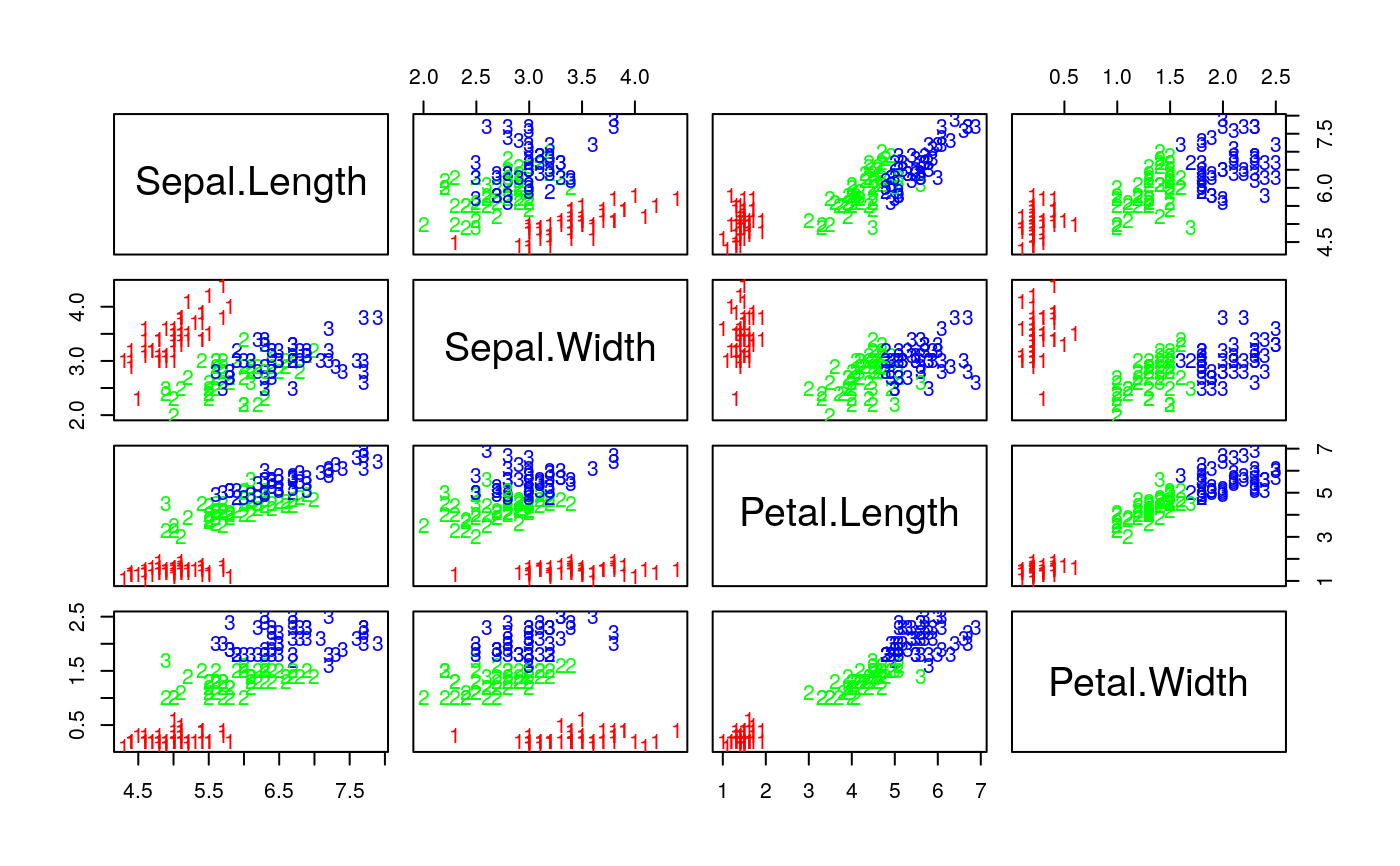
data(iris) cl <- specClust(iris[,1:4], 3, nn=5) pcol <- as.character(as.numeric(iris$Species)) pairs(iris[1:4], pch = pcol, col = c("green", "red", "blue")[cl$cluster])#> #> 1 2 3 #> setosa 0 50 0 #> versicolor 48 0 2 #> virginica 4 0 46