drawing phylogenetic tree from phylo object
Usage
ggevonet(tr, mapping = NULL, layout = "slanted", mrsd = NULL,
as.Date = FALSE, yscale = "none", yscale_mapping = NULL,
ladderize = FALSE, right = FALSE, branch.length = "branch.length",
ndigits = NULL, min_crossing = TRUE, ...)Arguments
- tr
a evonet object
- mapping
aes mapping
- layout
one of 'rectangular', 'slanted'
- mrsd
most recent sampling date
- as.Date
logical whether using Date class in time tree
- yscale
y scale
- yscale_mapping
yscale mapping for category variable
- ladderize
logical
- right
logical
- branch.length
variable for scaling branch, if 'none' draw cladogram
- ndigits
number of digits to round numerical annotation variable
- min_crossing
logical, rotate clades to minimize crossings
- ...
additional parameter
Examples
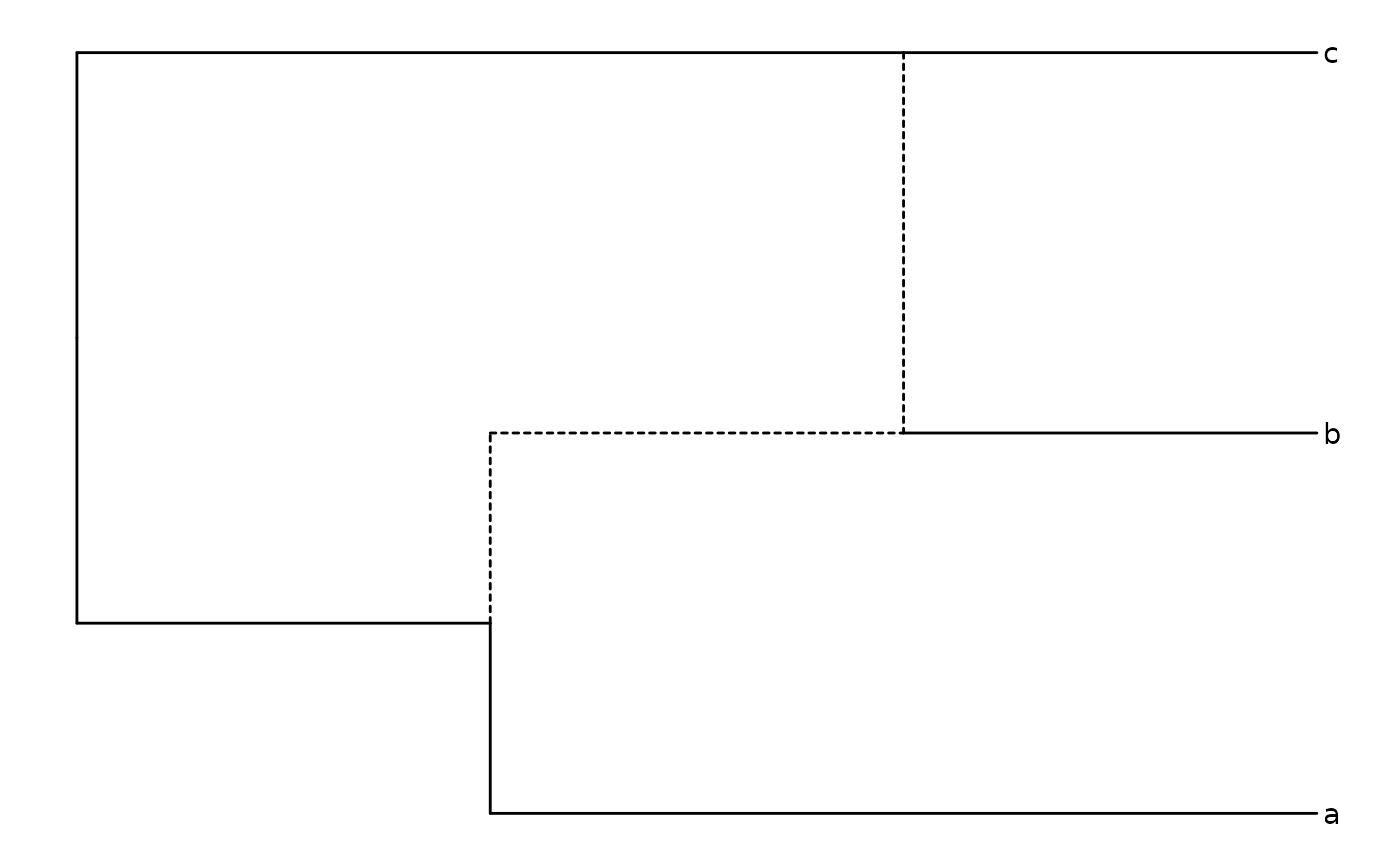
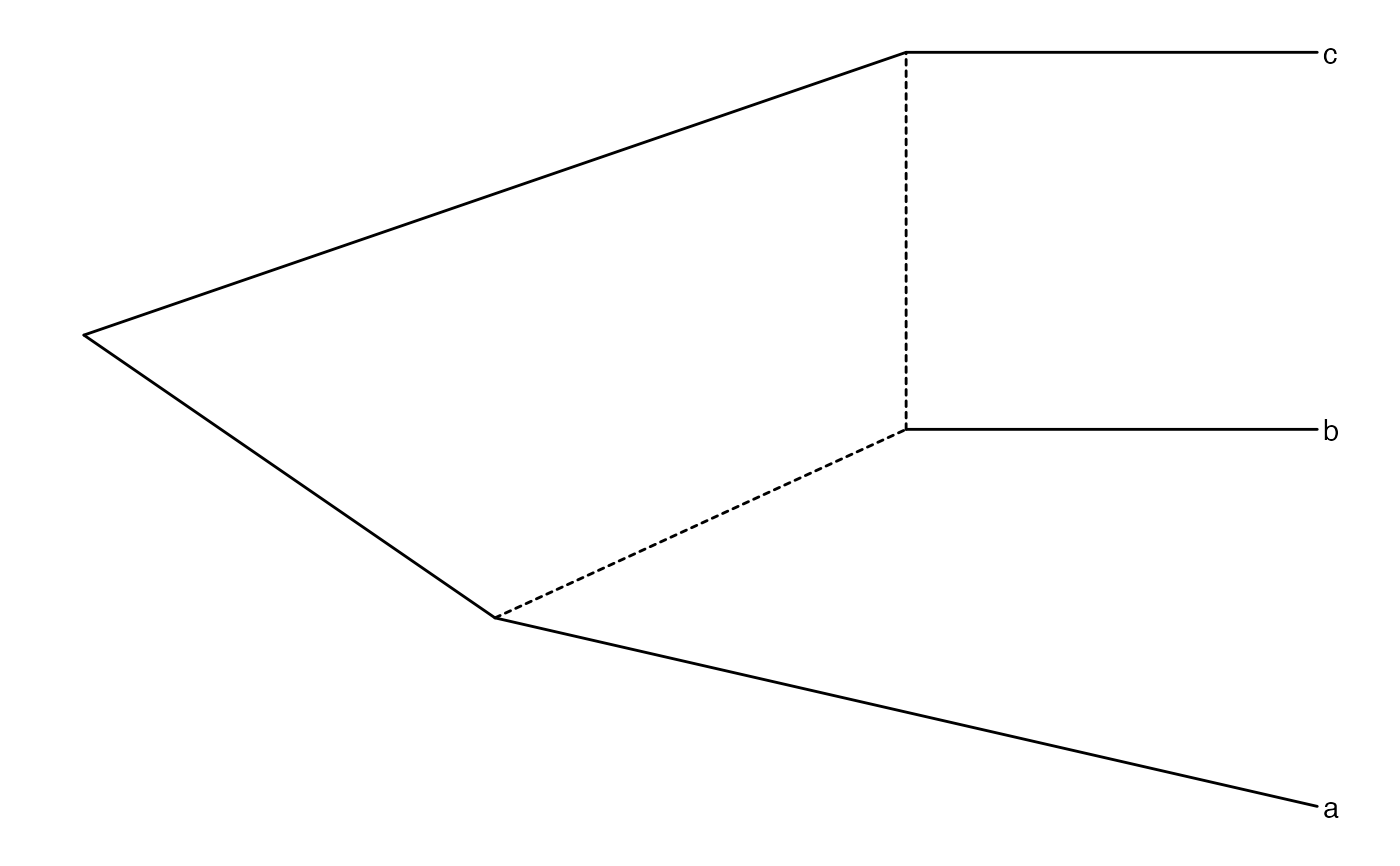
(enet <- ape::read.evonet(text='((a:2,(b:1)#H1:1):1,(#H1,c:1):2);'))
#>
#> Evolutionary network with 1 reticulation
#>
#> --- Base tree ---
#> Phylogenetic tree with 3 tips and 4 internal nodes.
#>
#> Tip labels:
#> a, b, c
#> Node labels:
#> , , #H1,
#>
#> Rooted; includes branch length(s).
ggevonet(enet) + geom_tiplab()
#> Warning: `aes_string()` was deprecated in ggplot2 3.0.0.
#> ℹ Please use tidy evaluation idioms with `aes()`.
#> ℹ See also `vignette("ggplot2-in-packages")` for more information.
#> ℹ The deprecated feature was likely used in the ggtree package.
#> Please report the issue at <https://github.com/YuLab-SMU/ggtree/issues>.
#> Warning: `aes_()` was deprecated in ggplot2 3.0.0.
#> ℹ Please use tidy evaluation idioms with `aes()`
#> ℹ The deprecated feature was likely used in the ggtree package.
#> Please report the issue at <https://github.com/YuLab-SMU/ggtree/issues>.
 ggevonet(enet, layout = "rectangular") + geom_tiplab()
ggevonet(enet, layout = "rectangular") + geom_tiplab()