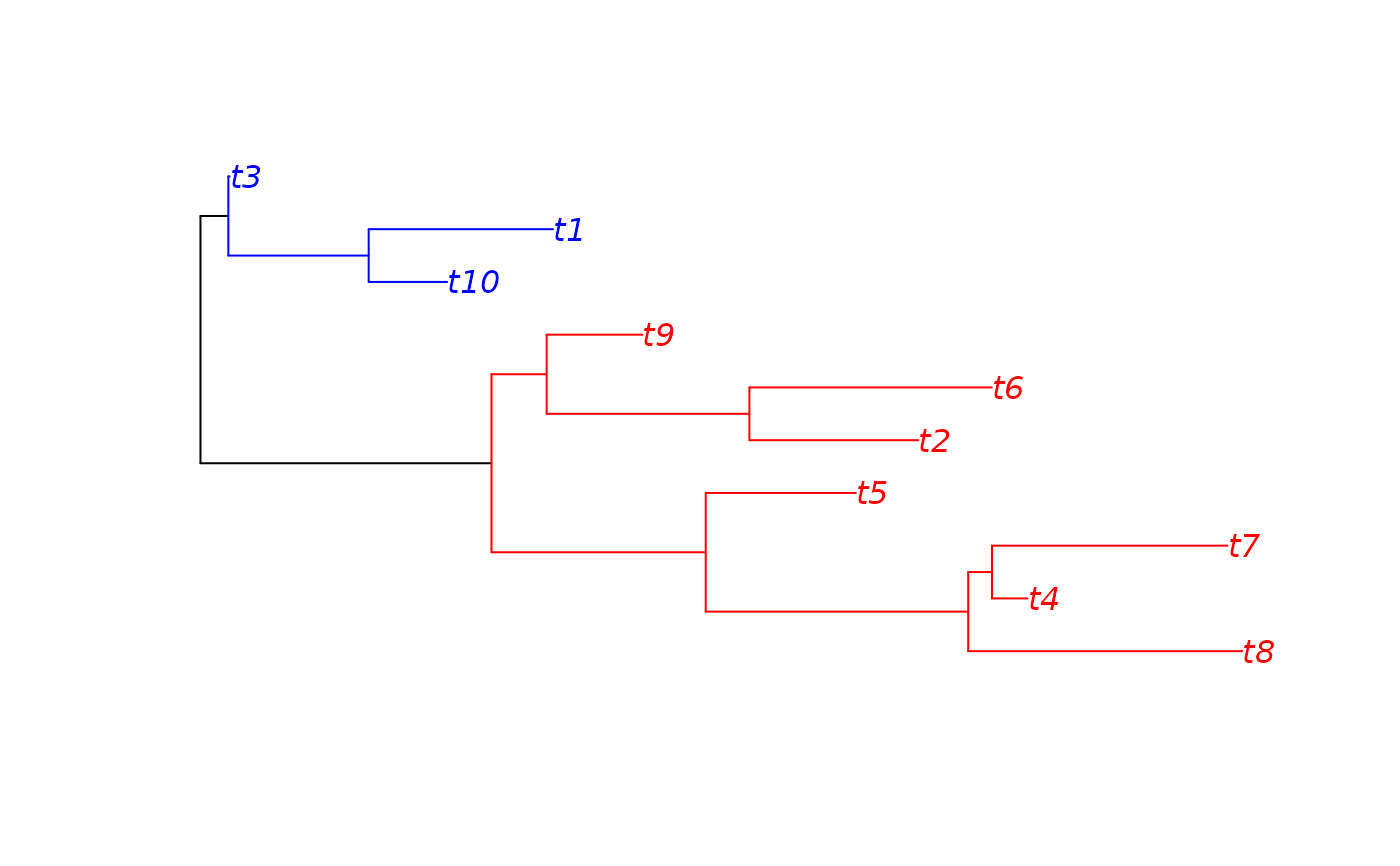
cladePar can help you coloring (choosing edge width/type) of clades.
Usage
cladePar(tree, node, edge.color = "red", tip.color = edge.color,
edge.width = 1, edge.lty = "solid", x = NULL, plot = FALSE, ...)Arguments
- tree
an object of class phylo.
- node
the node which is the common ancestor of the clade.
- edge.color
see plot.phylo.
- tip.color
see plot.phylo.
- edge.width
see plot.phylo.
- edge.lty
see plot.phylo.
- x
the result of a previous call to cladeInfo.
- plot
logical, if TRUE the tree is plotted.
- ...
Further arguments passed to or from other methods.
Author
Klaus Schliep klaus.schliep@gmail.com
Examples
tree <- rtree(10)
plot(tree)
nodelabels()
 x <- cladePar(tree, 12)
cladePar(tree, 18, "blue", "blue", x=x, plot=TRUE)
x <- cladePar(tree, 12)
cladePar(tree, 18, "blue", "blue", x=x, plot=TRUE)