drawing phylogenetic tree from phylo object
Usage
ggsplitnet(tr, mapping = NULL, layout = "slanted", mrsd = NULL,
as.Date = FALSE, yscale = "none", yscale_mapping = NULL,
ladderize = FALSE, right = FALSE, branch.length = "branch.length",
ndigits = NULL, angle = 0, ...)Arguments
- tr
a networx object
- mapping
aes mapping
- layout
so far only 'slanted' is supported.
- mrsd
most recent sampling date
- as.Date
logical whether using Date class in time tree
- yscale
y scale
- yscale_mapping
yscale mapping for category variable
- ladderize
logical
- right
logical
- branch.length
variable for scaling branch, if 'none' draw cladogram
- ndigits
number of digits to round numerical annotation variable
- angle
rotate the plot.
- ...
additional parameter
References
Schliep, K., Potts, A. J., Morrison, D. A. and Grimm, G. W. (2017), Intertwining phylogenetic trees and networks. Methods Ecol Evol. 8, 1212–1220. doi:10.1111/2041-210X.12760
Dress, A.W.M. and Huson, D.H. (2004) Constructing Splits Graphs IEEE/ACM Transactions on Computational Biology and Bioinformatics (TCBB), 1(3), 109–115
Bagci, C., Bryant, D., Cetinkaya, B. and Huson, D.H. (2021), Microbial Phylogenetic Context Using Phylogenetic Outlines. Genome Biology and Evolution. 13(9), evab213
Potts, A.J. and Hedderson, T.A. and Grimm, G.W. (2013), Constructing Phylogenies in the Presence Of Intra-Individual Site Polymorphisms (2ISPs) with a Focus on the Nuclear Ribosomal Cistron, Systematic Biology. 63(1), 1–16
Examples
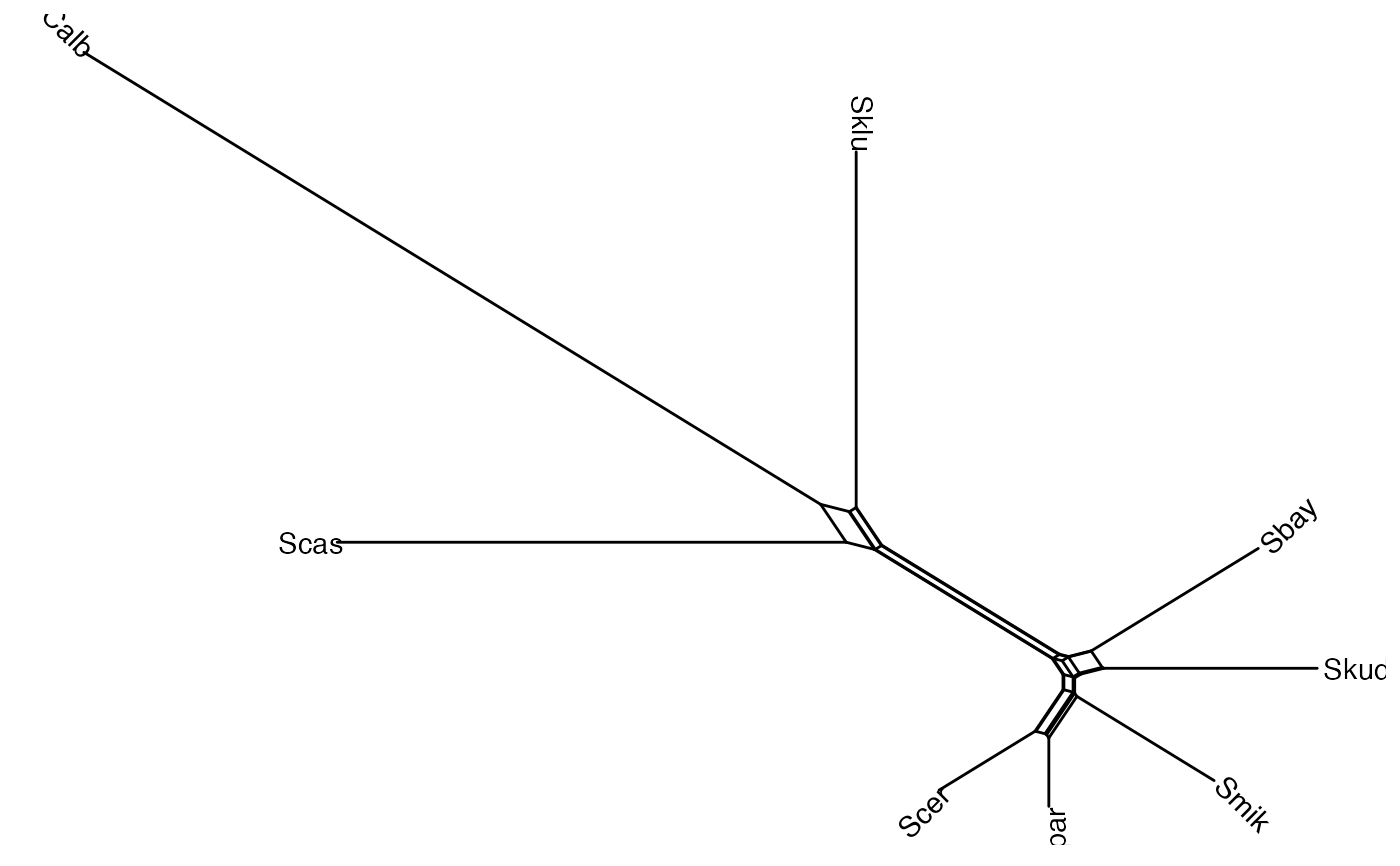
data(yeast, package='phangorn')
dm <- phangorn::dist.ml(yeast)
nnet <- phangorn::neighborNet(dm)
ggsplitnet(nnet) + geom_tiplab2()
 library(phangorn)
#> Loading required package: ape
#>
#> Attaching package: ‘ape’
#> The following object is masked from ‘package:ggtree’:
#>
#> rotate
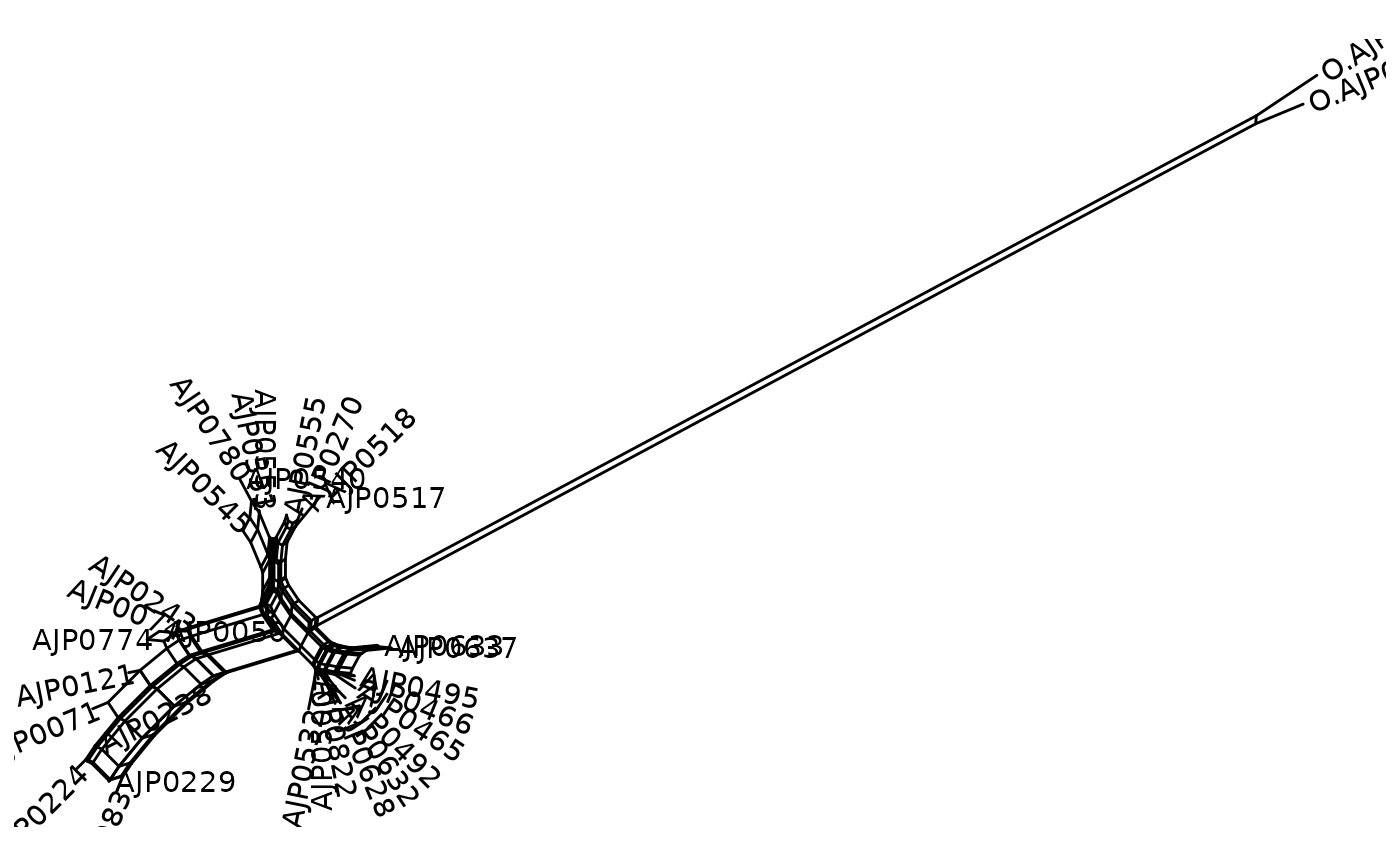
fdir <- system.file("extdata/examples", package = "tanggle")
nymania <- read.phyDat(file.path(fdir,
"Nymania.capensis.ITS.alignment.fasta"), format="fasta")
nnet <- neighborNet(dist.p(nymania))
ggsplitnet(nnet) + geom_tiplab2()
library(phangorn)
#> Loading required package: ape
#>
#> Attaching package: ‘ape’
#> The following object is masked from ‘package:ggtree’:
#>
#> rotate
fdir <- system.file("extdata/examples", package = "tanggle")
nymania <- read.phyDat(file.path(fdir,
"Nymania.capensis.ITS.alignment.fasta"), format="fasta")
nnet <- neighborNet(dist.p(nymania))
ggsplitnet(nnet) + geom_tiplab2()