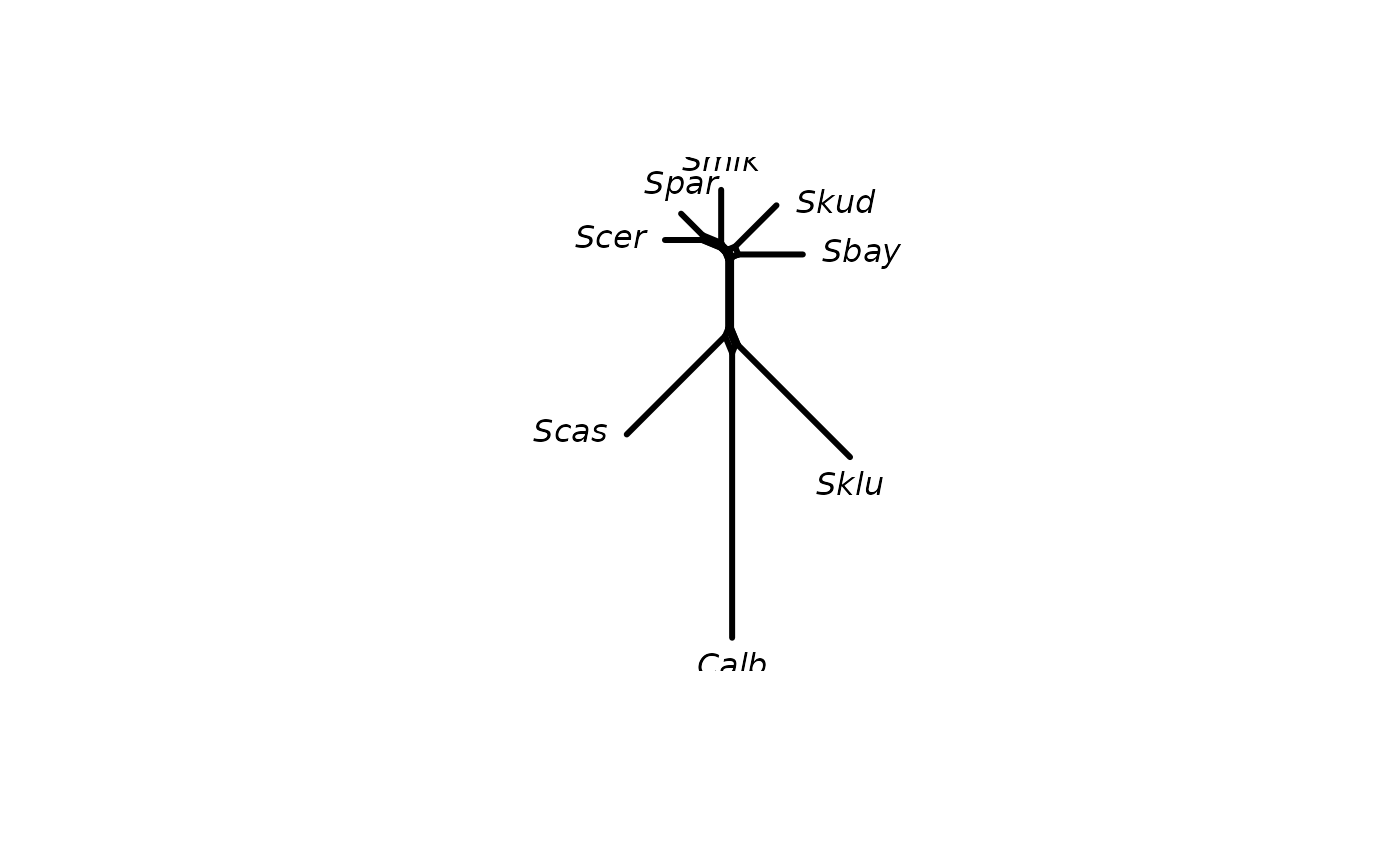
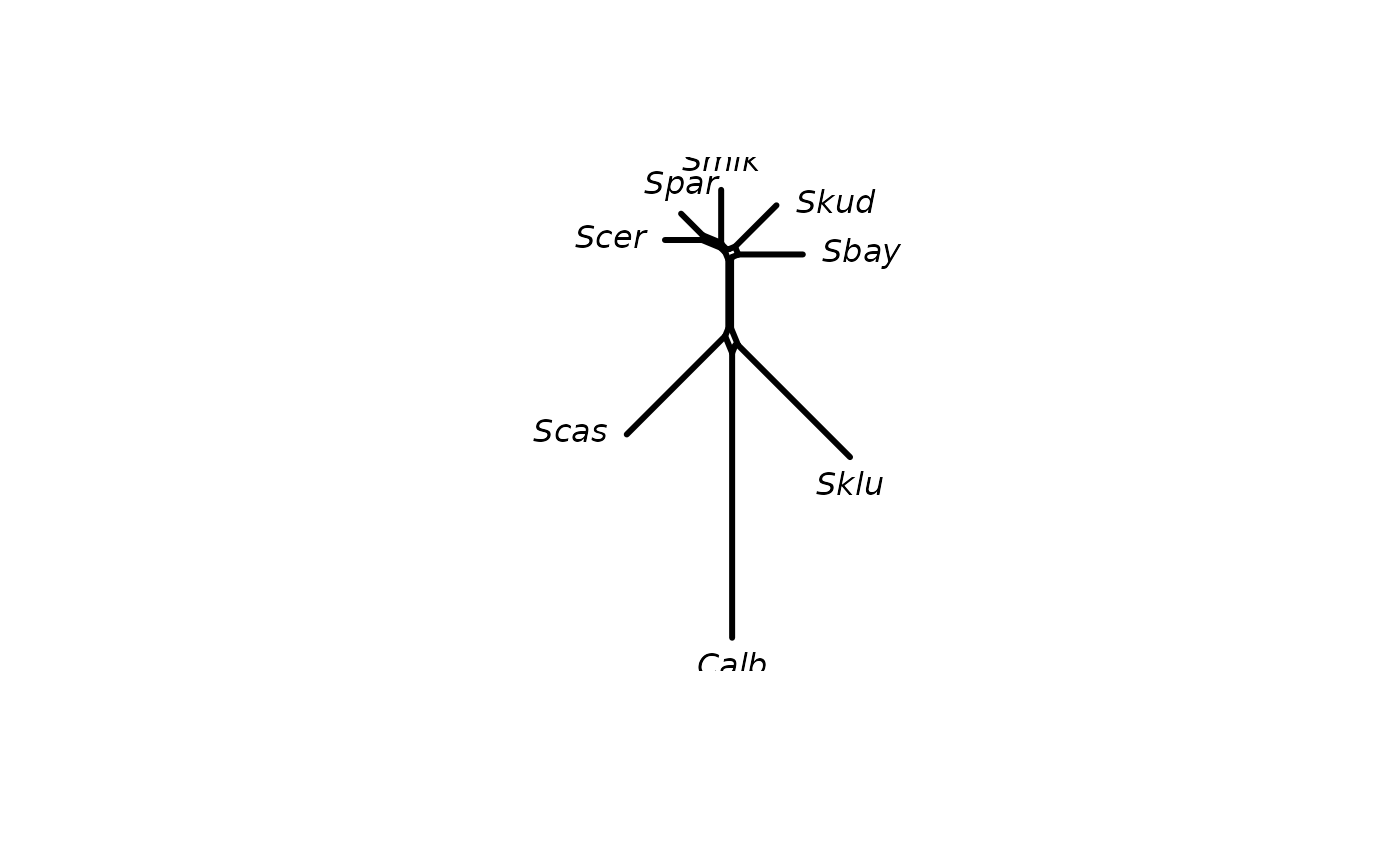
Computes a neighborNet, i.e. an object of class networx from a
distance matrix.
Details
neighborNet is still experimental. The cyclic ordering sometimes
differ from the SplitsTree implementation, the ord argument can be
used to enforce a certain circular ordering.
References
Bryant, D. & Moulton, V. (2004) Neighbor-Net: An Agglomerative Method for the Construction of Phylogenetic Networks. Molecular Biology and Evolution, 21, 255-265
Bryant, D., & Huson, D. H. (2023). NeighborNet: improved algorithms and implementation. Frontiers in bioinformatics, 3, 1178600.
Author
Klaus Schliep klaus.schliep@gmail.com