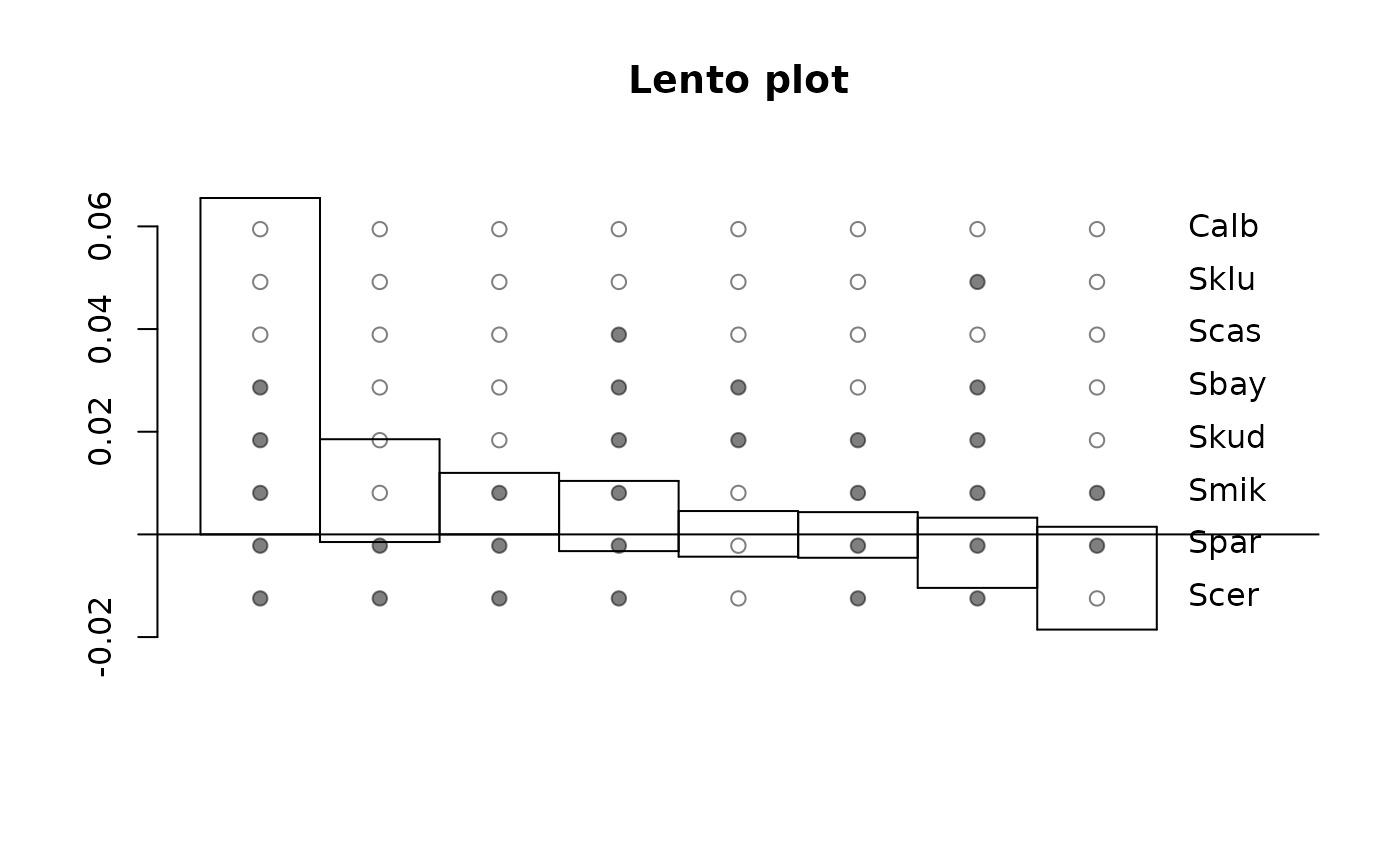
Distance Hadamard produces spectra of splits from a distance matrix.
Value
distanceHadamard returns a matrix. The first column contains
the distance spectra, the second one the edge-spectra. If eps is positive an
object of with all splits greater eps is returned.
References
Hendy, M. D. and Penny, D. (1993). Spectral Analysis of Phylogenetic Data. Journal of Classification, 10, 5-24.
Author
Klaus Schliep klaus.schliep@gmail.com, Tim White
Examples
data(yeast)
dm <- dist.hamming(yeast)
dm <- as.matrix(dm)
fit <- distanceHadamard(dm)
lento(fit)
 plot(as.networx(fit))
plot(as.networx(fit))