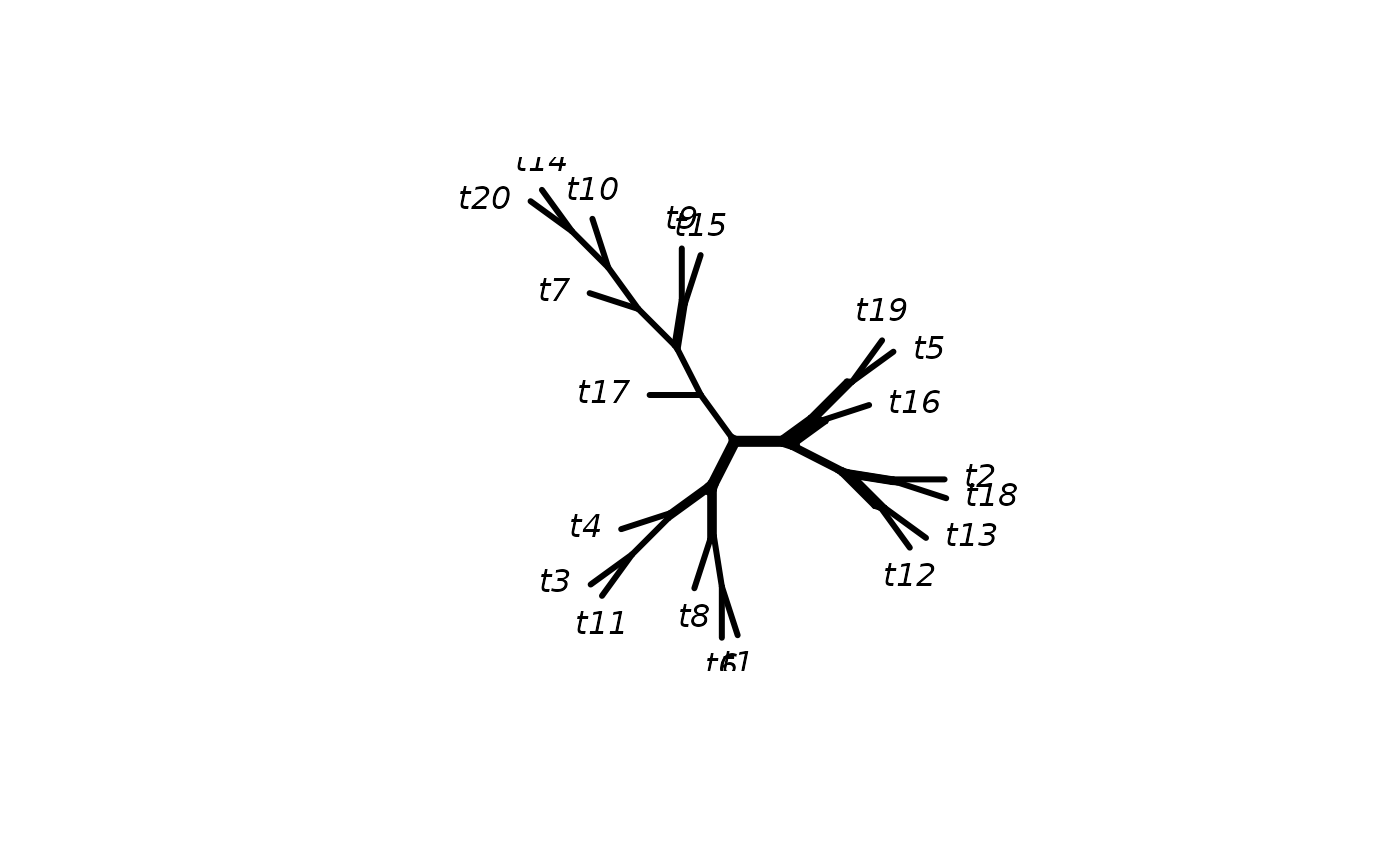
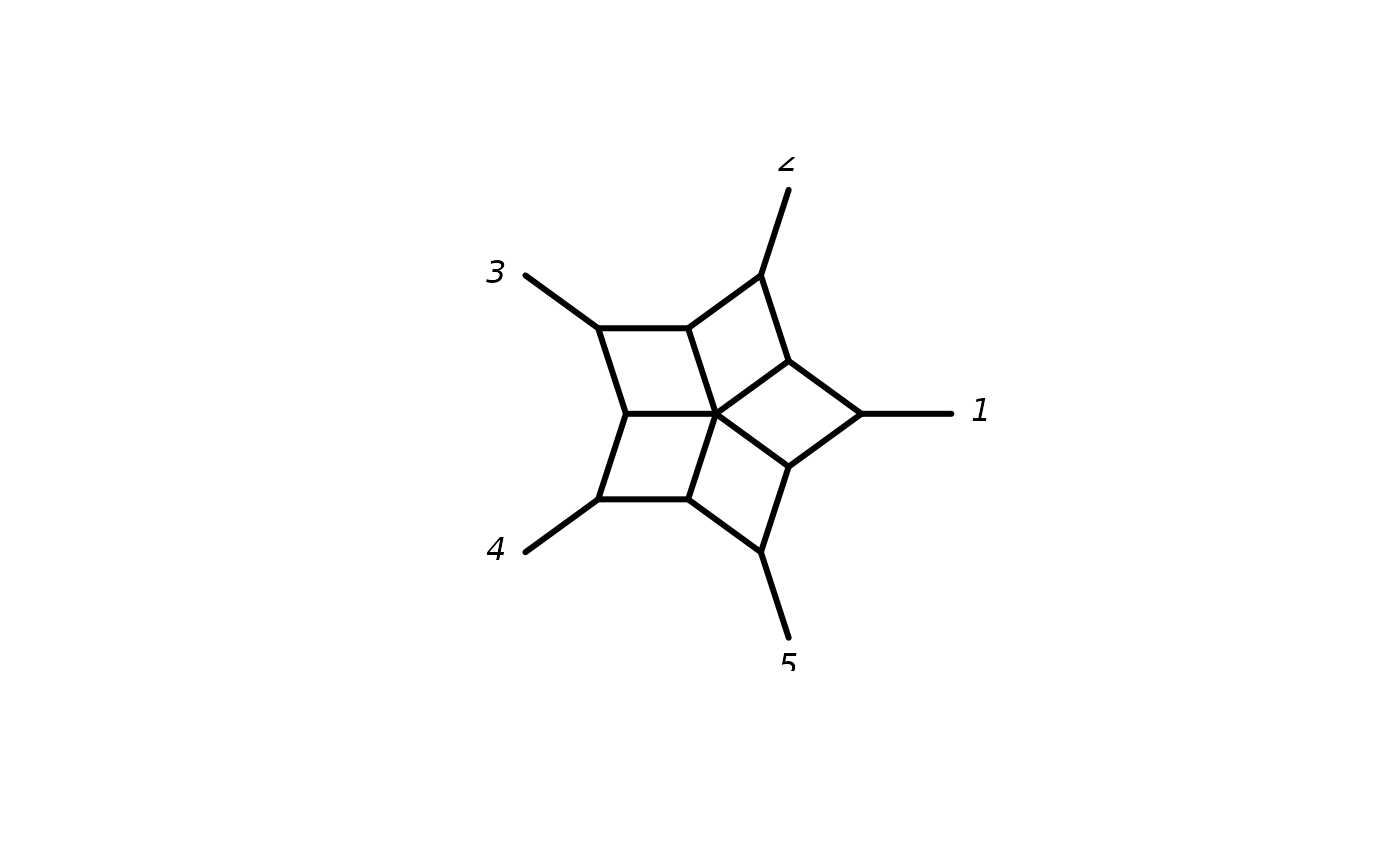
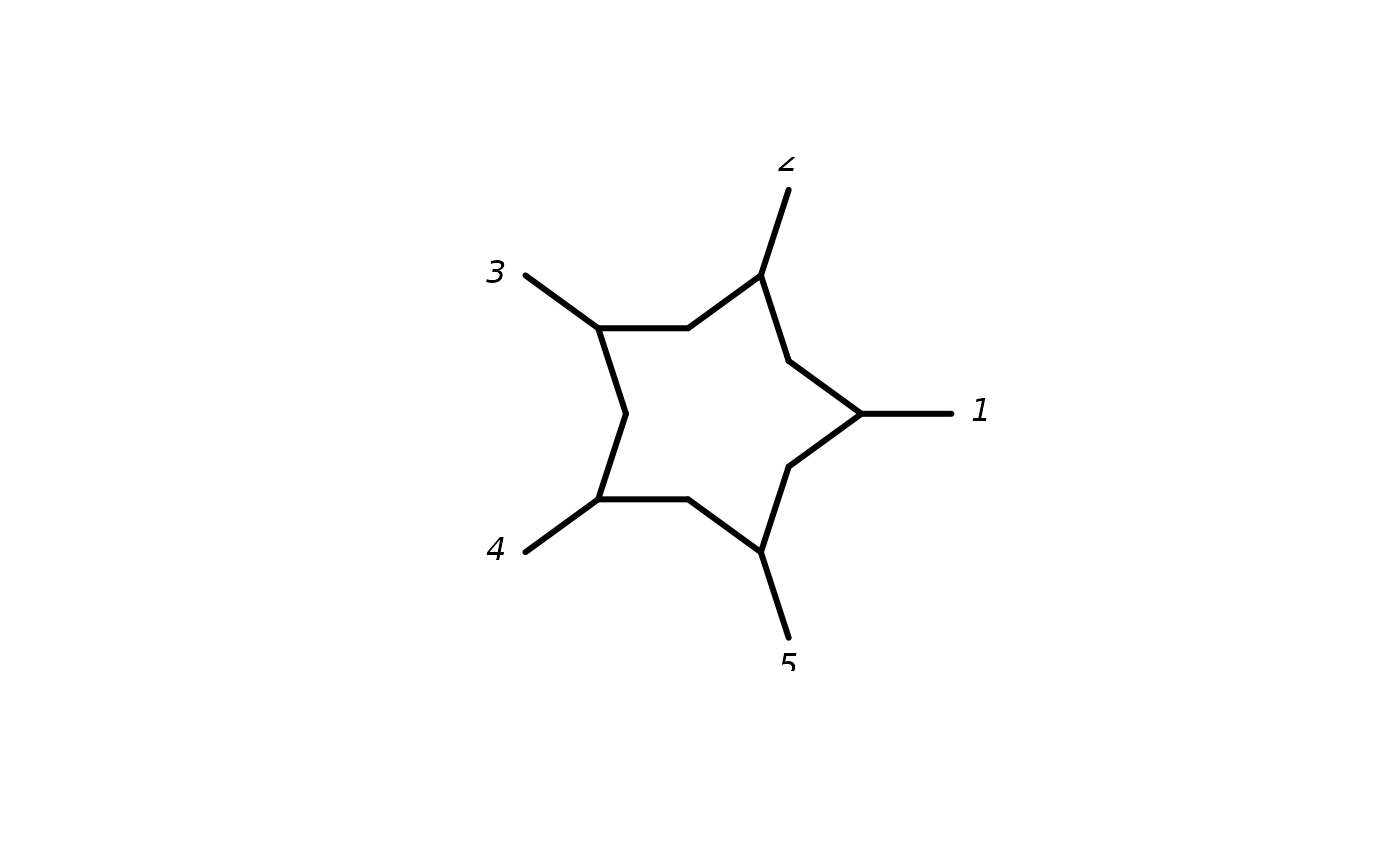
as.networx convert splits objects into a networx
object. And most important there exists a generic plot function to
plot phylogenetic network or split graphs.
Usage
as.networx(x, ...)
# S3 method for class 'splits'
as.networx(x, planar = FALSE, coord = "none", ...)
# S3 method for class 'phylo'
as.networx(x, ...)Details
A networx object hold the information for a phylogenetic
network and extends the phylo object. Therefore some generic function
for phylo objects will also work for networx objects. The
argument planar = TRUE will create a planar split graph based on a
cyclic ordering. These objects can be nicely plotted in "2D".
The argument "coord" allows to create coordinates, with options are "none",
"equal angle", "3D", "2D" and "outline" (Bagci et al. 2021).
References
Schliep, K., Potts, A. J., Morrison, D. A. and Grimm, G. W. (2017), Intertwining phylogenetic trees and networks. Methods Ecol Evol. 8, 1212–1220. doi:10.1111/2041-210X.12760
Bagci, C., Bryant, D., Cetinkaya, B. and Huson, D.H. (2021), Microbial Phylogenetic Context Using Phylogenetic Outlines. Genome Biology and Evolution. Volume 13. Issue 9. evab213
Author
Klaus Schliep klaus.schliep@gmail.com