
Function to import and export splits and networks
Source:R/read.nexus.splits.R
read.nexus.splits.Rdread.nexus.splits, write.nexus.splits,
read.nexus.networx, write.nexus.networx
can be used to import and export splits and networks with nexus format
and allow to exchange these object with other software like SplitsTree.
write.splits returns a human readable output.
Usage
read.nexus.splits(file)
write.nexus.splits(obj, file = "", weights = NULL, taxa = TRUE,
append = FALSE)
write.nexus.networx(obj, file = "", taxa = TRUE, splits = TRUE,
append = FALSE)
read.nexus.networx(file, splits = TRUE)
write.splits(x, file = "", zero.print = ".", one.print = "|",
print.labels = TRUE, ...)Arguments
- file
a file name.
- obj
An object of class splits.
- weights
Edge weights.
- taxa
logical. If TRUE a taxa block is added
- append
logical. If TRUE the nexus blocks will be added to a file.
- splits
logical. If TRUE the nexus blocks will be added to a file.
- x
An object of class splits.
- zero.print
character which should be printed for zeros.
- one.print
character which should be printed for ones.
- print.labels
logical. If TRUE labels are printed.
- ...
Further arguments passed to or from other methods.
- labels
names of taxa.
Value
write.nexus.splits and write.nexus.networx write out
the splits and networx object to read with
other software like SplitsTree.
read.nexus.splits and read.nexus.networx return an
splits and networx object.
Note
read.nexus.splits reads in the splits block of a nexus file. It
assumes that different co-variables are tab delimited and the bipartition
are separated with white-space. Comments in square brackets are ignored.
Author
Klaus Schliep klaus.schliep@gmail.com
Examples
(sp <- as.splits(rtree(5)))
#> t4 t1 t2 t3 t5
#> [1,] | . . . .
#> [2,] . | . . .
#> [3,] . . | . .
#> [4,] . . . | .
#> [5,] . . . . |
#> [6,] | | | | |
#> [7,] . | | | |
#> [8,] . . | | |
#> [9,] . . | | .
write.nexus.splits(sp)
#> #NEXUS
#>
#> [Splits block for Spectronet or SplitsTree]
#> [generated by phangorn 2.12.1.3 ]
#>
#> BEGIN TAXA;
#> DIMENSIONS ntax=5;
#> TAXLABELS t4 t1 t2 t3 t5 ;
#> END;
#>
#> BEGIN SPLITS;
#> DIMENSIONS ntax=5 nsplits=8;
#> FORMAT labels=left weights=yes confidences=no intervals=no;
#> MATRIX
#> 1 0.234915791545063 1,
#> 2 0.264620627742261 1 3 4 5,
#> 3 0.0627861295361072 1 2 4 5,
#> 4 0.397706091869622 1 2 3 5,
#> 5 0.490028424886987 1 2 3 4,
#> 6 0.021011842880398 1,
#> 7 0.691801619483158 1 2,
#> 8 0.0346693911124021 1 2 5,
#> ;
#> END;
spl <- allCircularSplits(5)
plot(as.networx(spl))
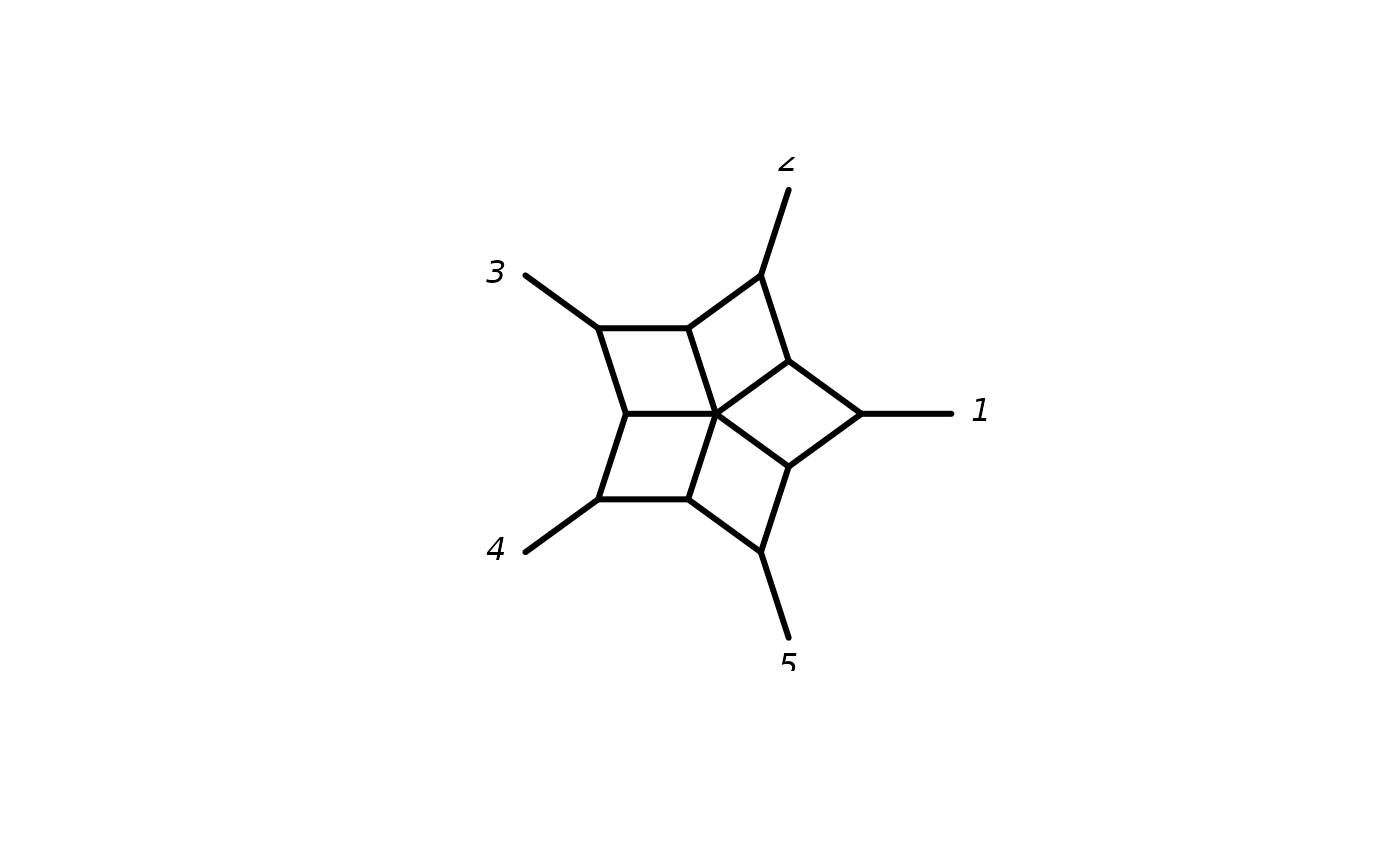 write.splits(spl, print.labels = FALSE)
#>
#>
#> |....
#>
#> .|...
#>
#> ..|..
#>
#> ...|.
#>
#> ....|
#>
#> ||...
#>
#> .||..
#>
#> ..||.
#>
#> ...||
#>
#> |...|
write.splits(spl, print.labels = FALSE)
#>
#>
#> |....
#>
#> .|...
#>
#> ..|..
#>
#> ...|.
#>
#> ....|
#>
#> ||...
#>
#> .||..
#>
#> ..||.
#>
#> ...||
#>
#> |...|