nnls.tree estimates the branch length using non-negative least
squares given a tree and a distance matrix. designTree and
designSplits compute design matrices for the estimation of edge
length of (phylogenetic) trees using linear models. For larger trees a
sparse design matrix can save a lot of memory. designTree also
computes a contrast matrix if the method is "rooted".
Usage
designTree(tree, method = "unrooted", sparse = FALSE, tip.dates = NULL,
calibration = NULL, ...)
nnls.tree(dm, tree, method = c("unrooted", "ultrametric", "tipdated"),
rooted = NULL, trace = 1, weight = NULL, balanced = FALSE,
tip.dates = NULL, calibration = NULL)
nnls.phylo(x, dm, method = "unrooted", trace = 0, ...)
nnls.splits(x, dm, trace = 0, eps = 1e-08)
nnls.networx(x, dm, eps = 1e-08)
designSplits(x, splits = "all", ...)Arguments
- tree
an object of class
phylo- method
compute an "unrooted", "ultrametric" or "tipdated" tree.
- sparse
return a sparse design matrix.
- tip.dates
a named vector of sampling times associated to the tips of the tree.
- calibration
a named vector of calibration times associated to nodes of the tree.
- ...
further arguments, passed to other methods.
- dm
a distance matrix.
- rooted
compute a "ultrametric" or "unrooted" tree (better use method).
- trace
defines how much information is printed during optimization.
- weight
vector of weights to be used in the fitting process. Weighted least squares is used with weights w, i.e., sum(w * e^2) is minimized.
- balanced
use weights as in balanced fastME
- x
number of taxa.
- eps
minimum edge length (default s 1e-8).
- splits
one of "all", "star".
Value
nnls.tree return a tree, i.e. an object of class
phylo. designTree and designSplits a matrix, possibly
sparse.
Author
Klaus Schliep klaus.schliep@gmail.com
Examples
example(NJ)
#>
#> NJ> data(Laurasiatherian)
#>
#> NJ> dm <- dist.ml(Laurasiatherian)
#>
#> NJ> tree <- NJ(dm)
#>
#> NJ> plot(tree)
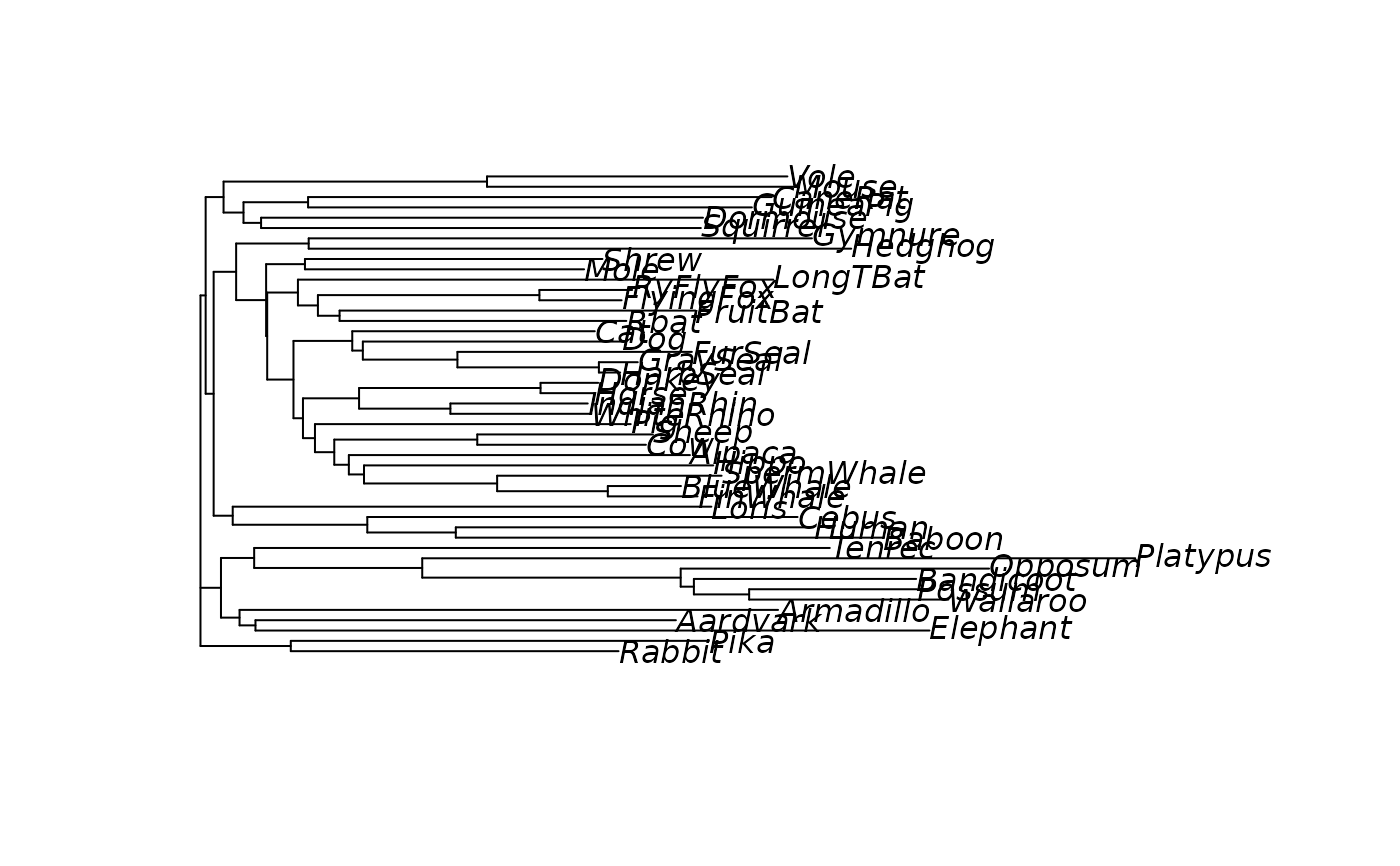 dm <- as.matrix(dm)
y <- dm[lower.tri(dm)]
X <- designTree(tree)
lm(y~X-1)
#>
#> Call:
#> lm(formula = y ~ X - 1)
#>
#> Coefficients:
#> X89<->36 X89<->37 X77<->35 X77<->38 X73<->33 X73<->34
#> 0.0499157 0.0491439 0.0724626 0.0755455 0.0712616 0.0725432
#> X65<->73 X65<->77 X51<->65 X51<->89 X75<->10 X75<->11
#> 0.0034694 0.0099328 0.0029426 0.0432799 0.0885305 0.0820387
#> X62<->12 X62<->13 X86<->15 X86<->16 X67<->14 X67<->17
#> 0.0461128 0.0479378 0.0134025 0.0151296 0.0467324 0.0580930
#> X63<->67 X63<->86 X59<->63 X59<->18 X88<->45 X88<->47
#> 0.0035646 0.0360953 0.0032091 0.0775894 0.0034977 0.0063178
#> X80<->88 X80<->46 X74<->80 X74<->44 X72<->74 X72<->43
#> 0.0231875 0.0381300 0.0154870 0.0421651 0.0015801 0.0392256
#> X85<->19 X85<->20 X79<->21 X79<->22 X71<->79 X71<->85
#> 0.0088520 0.0092959 0.0229321 0.0224528 0.0145614 0.0299323
#> X81<->25 X81<->26 X87<->28 X87<->29 X82<->87 X82<->30
#> 0.0274720 0.0286839 0.0148471 0.0117270 0.0181692 0.0364881
#> X70<->82 X70<->27 X68<->70 X68<->24 X66<->68 X66<->81
#> 0.0219311 0.0563100 0.0033377 0.0569549 0.0008004 0.0221615
#> X61<->66 X61<->23 X60<->61 X60<->71 X58<->60 X58<->72
#> 0.0021831 0.0502453 0.0042762 0.0102058 0.0020978 0.0117101
#> X57<->58 X57<->59 X56<->57 X56<->62 X53<->56 X53<->75
#> 0.0013537 0.0043899 0.0011632 0.0063020 0.0070446 0.0117666
#> X84<->39 X84<->40 X78<->84 X78<->42 X52<->78 X52<->41
#> 0.0696111 0.0583581 0.0144780 0.0701359 0.0225117 0.0792075
#> X50<->52 X50<->53 X49<->50 X49<->51 X92<->2 X92<->3
#> 0.0008563 0.0018880 0.0036910 0.0033483 0.0324915 0.0274029
#> X91<->92 X91<->4 X90<->91 X90<->5 X83<->90 X83<->1
#> 0.0090135 0.0362511 0.0024394 0.0505988 0.0425011 0.1170301
#> X69<->83 X69<->9 X64<->7 X64<->8 X55<->64 X55<->6
#> 0.0289009 0.0967237 0.1091965 0.0693124 0.0029756 0.0874519
#> X54<->55 X54<->69 X76<->31 X76<->32 X48<->76 X48<->54
#> 0.0027613 0.0065884 0.0534621 0.0681912 0.0151764 0.0010911
#> X48<->49
#> -0.0018629
#>
# avoids negative edge weights
tree2 <- nnls.tree(dm, tree)
dm <- as.matrix(dm)
y <- dm[lower.tri(dm)]
X <- designTree(tree)
lm(y~X-1)
#>
#> Call:
#> lm(formula = y ~ X - 1)
#>
#> Coefficients:
#> X89<->36 X89<->37 X77<->35 X77<->38 X73<->33 X73<->34
#> 0.0499157 0.0491439 0.0724626 0.0755455 0.0712616 0.0725432
#> X65<->73 X65<->77 X51<->65 X51<->89 X75<->10 X75<->11
#> 0.0034694 0.0099328 0.0029426 0.0432799 0.0885305 0.0820387
#> X62<->12 X62<->13 X86<->15 X86<->16 X67<->14 X67<->17
#> 0.0461128 0.0479378 0.0134025 0.0151296 0.0467324 0.0580930
#> X63<->67 X63<->86 X59<->63 X59<->18 X88<->45 X88<->47
#> 0.0035646 0.0360953 0.0032091 0.0775894 0.0034977 0.0063178
#> X80<->88 X80<->46 X74<->80 X74<->44 X72<->74 X72<->43
#> 0.0231875 0.0381300 0.0154870 0.0421651 0.0015801 0.0392256
#> X85<->19 X85<->20 X79<->21 X79<->22 X71<->79 X71<->85
#> 0.0088520 0.0092959 0.0229321 0.0224528 0.0145614 0.0299323
#> X81<->25 X81<->26 X87<->28 X87<->29 X82<->87 X82<->30
#> 0.0274720 0.0286839 0.0148471 0.0117270 0.0181692 0.0364881
#> X70<->82 X70<->27 X68<->70 X68<->24 X66<->68 X66<->81
#> 0.0219311 0.0563100 0.0033377 0.0569549 0.0008004 0.0221615
#> X61<->66 X61<->23 X60<->61 X60<->71 X58<->60 X58<->72
#> 0.0021831 0.0502453 0.0042762 0.0102058 0.0020978 0.0117101
#> X57<->58 X57<->59 X56<->57 X56<->62 X53<->56 X53<->75
#> 0.0013537 0.0043899 0.0011632 0.0063020 0.0070446 0.0117666
#> X84<->39 X84<->40 X78<->84 X78<->42 X52<->78 X52<->41
#> 0.0696111 0.0583581 0.0144780 0.0701359 0.0225117 0.0792075
#> X50<->52 X50<->53 X49<->50 X49<->51 X92<->2 X92<->3
#> 0.0008563 0.0018880 0.0036910 0.0033483 0.0324915 0.0274029
#> X91<->92 X91<->4 X90<->91 X90<->5 X83<->90 X83<->1
#> 0.0090135 0.0362511 0.0024394 0.0505988 0.0425011 0.1170301
#> X69<->83 X69<->9 X64<->7 X64<->8 X55<->64 X55<->6
#> 0.0289009 0.0967237 0.1091965 0.0693124 0.0029756 0.0874519
#> X54<->55 X54<->69 X76<->31 X76<->32 X48<->76 X48<->54
#> 0.0027613 0.0065884 0.0534621 0.0681912 0.0151764 0.0010911
#> X48<->49
#> -0.0018629
#>
# avoids negative edge weights
tree2 <- nnls.tree(dm, tree)
